Status: Confirmed ARS: Confirmed by ARS assay and 2D gel and identified by 9 genome-wide studies.
Genomic Location:
Chr3:74457-74677
Within tandem intergenic space between YCL026C-B and YCL026C-A.
• View at UCSC genome browser
• View on SGD Chromosome Features Map
• View at SGD gbrowser site
• View at Ensembl browser
DNA Sequence:
Origin Sequence Elements: ACS confirmed.
Helical Stability: Minimum -145.0 ΔG° (kcal/mol) at location 74617.
Time of Origin Replication (Trep):
Raghuraman et al. (2001) — Trep: 10.8 min.
Yabuki et al. (2002) — Trep: 17.9 min.
Alvino et al. (2007) — Peak first observed at 10.0 min.
Origin activity in HU: Origin activity detected in HU — more details.
ARS at III-75 has unique ID: 71
Loading - Please wait...
ARS at III-75 has unique ID: 71
Studies that cloned this origin
Chr3:74457-74677
Studies that analyzed this origin by 2D gel
Chr3:72852-77073
Studies that detected this origin by chromatin immunoprecipitation (ChIP)
Chr3:71881-75850
Chr3:74200-75100 (orc2-1/wt peak ratio: 0.54)
Chr3:74555-74565
Chr3:74150-74750 (orc1-bah-delta/wt peak ratio: 0.29)
Studies that measured the replication time of this origin
Chr3:77855 (Trep: 10.8 min.) (Confidence: 9)
Chr3:74863 (Trep: 17.9 min.)
Chr3:72000 (Peak first observed at 10.0 min.)
Studies that measured the activity of this origin in hydroxyurea (HU)
Chr3:73500 (Activity detected in: wild-type, rad53)
Chr3:74457-74677 (Activity detected in: wild-type, rad9, rev3 rad30, eco1, ctf4, ddc1, rad24, pol2-12, elg1, tof1, mrc1-AQ, mrc1-AQ rad9, ctf8, ctf18, mrc1, dcc1, ctf18 rad9, mec1-100, rad53, mec1)
Studies that predicted the location of this origin
This origin has not been predicted by any curated study.
ARS at III-75 has unique ID: 71
Studies that confirmed an essential ACS element
| Theis et al. (1999): | A | T | T | T | A | T | A | T | T | T | A | T |
Studies that predicted an essential ACS element
| Xu et al. (2006): | T | G | T | A | T | T | T | A | T | A | T | T | T | A | T | T | T | A | G | T | A | C | T | T | C | T | T | G | A | G | T | T | T | A | ||||||||
| ACS LOGO: | 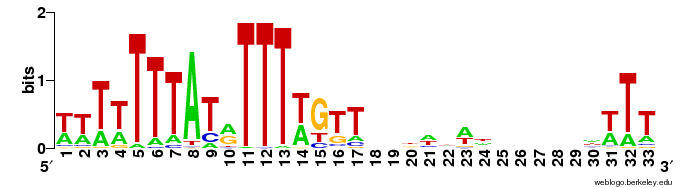 | |||||||||||||||||||||||||||||||||||||||||
ARS at III-75 has unique ID: 71
Alignments from the UCSC genome browser
No phylogenetic sequence conservation data.ARS at III-75 has unique ID: 71
These notes are manually curated. To submit notes for this replication origin site please contact us.
There are no notes entered for this replication origin site.
ARS at III-75 has unique ID: 71
Genome-wide studies that identified this origin
PubMed | PubMed Central | Nat. Cell Biol.
PubMed | PubMed Central | BMC Genomics
PubMed | PubMed Central | Mol. Cell. Biol.
PubMed | PubMed Central | PLoS Genet.
PubMed | PubMed Central | Nat. Struct. Mol. Biol.
PubMed | PubMed Central | Genes Dev.
PubMed | Nat. Struct. Mol. Biol.
Studies that cloned this origin
Studies that analyzed this origin by 2D gel
Studies that confirmed an essential ACS element
Studies that predicted an essential ACS element
PubMed | PubMed Central | BMC Genomics
ARS at III-75 has unique ID: 71