Status: Confirmed ARS: Confirmed by ARS assay and identified by 8 genome-wide studies.
Genomic Location:
Chr10:298471-298952
Within convergent intergenic space between YJL076W and YJL074C.
• View at UCSC genome browser
• View on SGD Chromosome Features Map
• View at SGD gbrowser site
• View at Ensembl browser
DNA Sequence:
Origin Sequence Elements: ACS predicted.
Helical Stability: Minimum -145.4 ΔG° (kcal/mol) at location 298911.
Time of Origin Replication (Trep):
Raghuraman et al. (2001) — Trep: 33.2 min.
Alvino et al. (2007) — Peak first observed at 15.0 min.
Origin activity in HU: Origin activity detected in HU — more details.
ARS at X-299 has unique ID: ARS1010
Loading - Please wait...
ARS at X-299 has unique ID: ARS1010
Studies that cloned this origin
Chr10:298471-298952
Studies that analyzed this origin by 2D gel
None curated.
Studies that detected this origin by chromatin immunoprecipitation (ChIP)
Chr10:298572-298853
Chr10:298095-299900
Chr10:298775-298785
Chr10:298700-299300 (orc1-bah-delta/wt peak ratio: 0.98)
Studies that measured the replication time of this origin
Chr10:304757 (Trep: 33.2 min.) (Confidence: 9)
Chr10:305890 (Peak first observed at 15.0 min.)
Studies that measured the activity of this origin in hydroxyurea (HU)
Chr10:300000 (Activity detected in: rad53)
Chr10:298471-298952 (Activity detected in: mrc1-AQ, mrc1-AQ rad9, ctf8, ctf18, mrc1, dcc1, ctf18 rad9, mec1-100, rad53, mec1)
Studies that predicted the location of this origin
This origin has not been predicted by any curated study.
ARS at X-299 has unique ID: ARS1010
Studies that confirmed an essential ACS element
Studies that predicted an essential ACS element
| Nieduszynski et al. (2006): | C | T | T | A | T | T | T | A | C | C | T | C | T | T | T | T | ||||||||||||||||||||||||||
| ACS LOGO: | 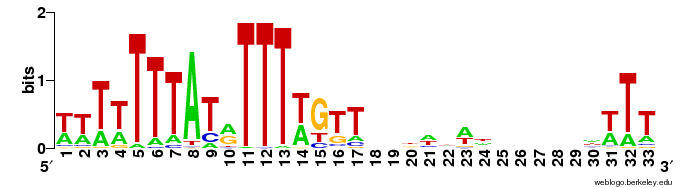 | |||||||||||||||||||||||||||||||||||||||||
ARS at X-299 has unique ID: ARS1010
Alignments from the UCSC genome browser
| Predicted ACS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| S. cerevisiae | A | A | T | C | T | T | C | A | T | G | T | A | A | A | C | C | A | A | T | T | C | T | T | A | T | T | T | A | C | C | T | C | T | T | T | T | C | C | C | A | G | C | A | A | G | C | G | G | G | C | A | A | C | T | T | C |
| S. kudriavzevii | A | T | T | T | T | T | C | A | T | G | T | A | A | A | C | T | A | A | T | - | - | T | T | A | T | T | C | A | T | C | T | C | T | A | C | G | A | T | A | A | T | C | A | - | - | T | G | A | T | C | A | A | - | T | T | T |
| S. mikatae | A | A | T | C | T | T | C | T | T | A | T | A | - | - | C | T | A | A | T | T | T | T | A | T | C | T | A | A | T | T | T | C | T | T | T | A | C | C | A | A | G | T | T | A | G | T | G | A | C | T | A | A | A | C | T | C |
| S. paradoxus | A | A | T | C | T | T | C | A | T | G | T | A | A | A | C | T | A | G | T | T | C | T | T | A | C | T | C | A | T | C | T | C | G | T | T | C | T | C | C | A | T | T | G | A | G | T | A | G | T | C | A | A | T | C | T | C |
| S. bayanus | A | T | T | T | C | T | C | A | T | G | T | A | A | A | C | T | A | A | T | T | C | T | - | A | T | T | T | A | T | C | T | C | C | T | T | C | A | A | T | A | T | T | A | A | A | T | A | A | G | T | A | G | A | T | T | T |
| * | * | : | * | * | : | * | : | * | * | : | : | * | : | * | : | * | : | * | : | * | * | : | : | * | * | : | : | : | * | : | : | * | : | : | * | |||||||||||||||||||||
ARS at X-299 has unique ID: ARS1010
These notes are manually curated. To submit notes for this replication origin site please contact us.
There are no notes entered for this replication origin site.
ARS at X-299 has unique ID: ARS1010
Genome-wide studies that identified this origin
PubMed | PubMed Central | Nat. Cell Biol.
PubMed | PubMed Central | BMC Genomics
PubMed | PubMed Central | Mol. Cell. Biol.
PubMed | PubMed Central | Nat. Struct. Mol. Biol.
PubMed | PubMed Central | Genes Dev.
PubMed | Nat. Struct. Mol. Biol.
Studies that cloned this origin
Studies that analyzed this origin by 2D gel
None identified.
Studies that confirmed an essential ACS element
None identified.
Studies that predicted an essential ACS element
PubMed | PubMed Central | Genes Dev.
ARS at X-299 has unique ID: ARS1010